Selecting representative tokens in R
25 Feb 2017 | all notes
alldata <- read.csv("Exp1CompRRVals.csv", stringsAsFactors=FALSE)
meaning.columns <- (ncol(alldata)-7) : ncol(alldata)
colnames(alldata)[meaning.columns] <- tolower(colnames(alldata)[meaning.columns])
meanings <- colnames(alldata)[meaning.columns]
cells <- NULL
cellsize <- 25
onewordperarea <- TRUE
selecttop <- function(ordereddata, meaning, n=cellsize) {
thismeaningcolumn <- match(meaning, colnames(ordereddata))
colnames(ordereddata)[thismeaningcolumn] <- "TargetMeaningRR"
# split matching and non-matching words into two data sets
correct <- subset(ordereddata, Meaning == meaning)
correctwords <- unique(correct$Word)
# only consider incorrect words which are of comparable length
correctwordlength <- range(sapply(correctwords, nchar))
incorrect <- subset(ordereddata, Meaning != meaning &
# only consider phonetic words which are incorrect in all languages
!(Word %in% correctwords) &
Length >= correctwordlength[1] &
Length <= correctwordlength[2])
# incorrectwords <- setdiff(unique(incorrect$Word), correctwords)
incorrectwords <- unique(incorrect$Word)
if (onewordperarea) {
# find n unique words which are also from n different glotto families
correctwordrows <- 1
incorrectwordrows <- 1
for (i in 2:n) {
# find next entry which is a new phonetic word and new glotto family
nextcorrectword <- subset(correct,
!(Word %in% correct$Word[correctwordrows]) &
!(Glotto %in% correct$Glotto[correctwordrows]))[1, c("Language", "Word")]
nextincorrectword <- subset(incorrect,
!(Word %in% incorrect$Word[incorrectwordrows]) &
!(Glotto %in% incorrect$Glotto[incorrectwordrows]))[1, c("Language", "Word")]
correctwordrows[i] <- which(correct$Language == nextcorrectword[[1]] & correct$Word == nextcorrectword[[2]])
incorrectwordrows[i] <- which(incorrect$Language == nextincorrectword[[1]] & incorrect$Word == nextincorrectword[[2]])
}
} else {
correctwordrows <- match(head(correctwords, n), correct$Word)
incorrectwordrows <- match(head(incorrectwords, n), incorrect$Word)
}
rbind(cbind(TargetMeaning=meaning, Matching=TRUE,
correct[correctwordrows, -setdiff(meaning.columns, thismeaningcolumn)]),
cbind(TargetMeaning=meaning, Matching=FALSE,
incorrect[incorrectwordrows, -setdiff(meaning.columns, thismeaningcolumn)]))
}
for (meaning in meanings) {
print(paste("Meaning", meaning))
data <- alldata[order(alldata[[meaning]], na.last=NA),]
mean <- mean(data[[meaning]])
median <- median(data[[meaning]])
print(paste("RR range from", data[1, meaning], "to", data[nrow(data), meaning],
"mean", mean, "median", median))
# simply reverse data frame order for highest
cells <- rbind(cells, cbind(RRclass="H", selecttop(data[nrow(data):1,], meaning)),
cbind(RRclass="L", selecttop(data, meaning)))
# select middle range:
# find range that is
highestlow <- max(subset(cells, TargetMeaning==meaning & RRclass=="L")$TargetMeaningRR)
lowesthigh <- min(subset(cells, TargetMeaning==meaning & RRclass=="H")$TargetMeaningRR)
untakenrangemean <- mean(c(highestlow, lowesthigh))
untakenrangemedian <- mean(sapply(c(highestlow, lowesthigh),
function(rr) which(data[[meaning]] == rr)[1]))
# order by absolute difference from mean or median (or untakenrangemean)
# data <- data[order(abs(data[[meaning]] - freerangemean)),]
# order by rank difference from median (nrow(data)/2) or untakenrangemedian
data <- data[order(abs(1:nrow(data) - untakenrangemedian)),]
cells <- rbind(cells, cbind(RRclass="M", selecttop(data, meaning)))
}
## [1] "Meaning bone"
## [1] "RR range from -2.36736894075312 to 1.26387134791426 mean -0.263666975626609 median -0.249721796954716"
## Warning in incorrectwordrows[i] <- which(incorrect$Language ==
## nextincorrectword[[1]] & : number of items to replace is not a multiple of
## replacement length
## [1] "Meaning breasts"
## [1] "RR range from -2.77010718884069 to 2.07297482061656 mean -0.335395309584245 median -0.352250311107756"
## [1] "Meaning dog"
## [1] "RR range from -2.46764753469791 to 1.75576955200288 mean -0.117006344871001 median -0.164695465190664"
## [1] "Meaning i"
## [1] "RR range from -5.11753380317081 to 1.51119327877114 mean -1.38262605491057 median -1.34345715548717"
## [1] "Meaning name"
## [1] "RR range from -3.41706133227802 to 1.51308486418524 mean -0.220013509567489 median -0.164209431621072"
## [1] "Meaning nose"
## [1] "RR range from -2.20471918813919 to 2.09233201159002 mean 0.12941416053513 median 0.13880894488938"
## [1] "Meaning tongue"
## [1] "RR range from -2.62921026006487 to 3.31591242468841 mean 0.164852303722952 median 0.142145751702432"
## [1] "Meaning we"
## [1] "RR range from -3.88497569181261 to 1.75264626903018 mean -0.350910710343393 median -0.352226981837328"
# safety check that all cells are completely filled
nrow(cells) == cellsize * length(meanings) * 6
## [1] TRUE
# and that we have the right number of unique (phonetic) words per target meaning
max(unique(xtabs(~ TargetMeaning + Word, data=cells))) == 1
## [1] TRUE
Make sure that the high/mid/low RR regions do not overlap – the six rows are the maximum/minimum target RR for the High, Mid and Low RR cells.
# reorder levels logically
cells$RRclass <- factor(cells$RRclass, levels = c("L", "M", "H"))
knitr::kable(sapply(as.character(unique(cells$TargetMeaning)),
function(m) sapply(levels(cells$RRclass),
function(rr) rev(range(subset(cells, RRclass==rr & TargetMeaning==m)$TargetMeaningRR)))))
| bone | breasts | dog | i | name | nose | tongue | we |
|---|---|---|---|---|---|---|---|
| -0.8395966 | -1.1633815 | -0.6913152 | -2.1246413 | -1.0060987 | -0.6881875 | -0.4422010 | -0.9260865 |
| -2.3673689 | -2.7701072 | -2.4676475 | -5.1175338 | -3.4170613 | -2.2047192 | -2.6292103 | -3.8849757 |
| -0.1991991 | -0.2843177 | -0.0985610 | -0.9038066 | -0.1125254 | 0.1878537 | 0.3098318 | -0.1771448 |
| -0.2346438 | -0.3230503 | -0.1422651 | -0.9881234 | -0.1615821 | 0.1496679 | 0.2672721 | -0.2329270 |
| 1.2638713 | 2.0729748 | 1.7557696 | 1.5111933 | 1.5130849 | 2.0923320 | 3.3159124 | 1.7526463 |
| 0.5698946 | 1.0207622 | 0.9114997 | 0.6486908 | 0.7162239 | 1.1808909 | 1.8623390 | 0.8233005 |
Check for words which made it into more than one cell due to overlap – overlap in ‘I’ can only be avoided by choosing the mid-RRs based on their rank within all of the RRs which aren’t covered by the high/low region.
cells[which(duplicated(cells[, c("TargetMeaning", "Language", "Word")])),
c("RRclass", "TargetMeaning", "Meaning", "Language", "Word", "TargetMeaningRR")]
## [1] RRclass TargetMeaning Meaning Language
## [5] Word TargetMeaningRR
## <0 rows> (or 0-length row.names)
if (onewordperarea)
cells[which(duplicated(cells[, c("RRclass", "TargetMeaning", "Matching", "Glotto")]))]
## data frame with 0 columns and 1200 rows
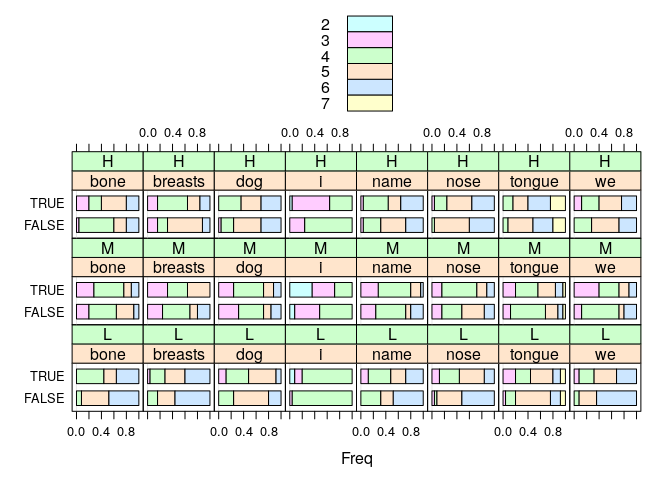
Compare the distribution of word lengths chosen for the matching/mismatching cells for every meaning.
x <- xtabs(~ Matching + TargetMeaning + RRclass + Length, data=cells)
lattice::barchart(x/array(rowSums(x, dim=3), dim=dim(x)), auto.key=TRUE)
Finally, look at the representedness of language areas in the sample.
perglotto <- table(cells$Glotto, cells$TargetMeaning)
knitr::kable(cbind(perglotto, total=rowSums(perglotto)))
| bone | breasts | dog | i | name | nose | tongue | we | total | |
|---|---|---|---|---|---|---|---|---|---|
| Abkhaz-Adyge | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 5 |
| Afro-Asiatic | 5 | 5 | 5 | 5 | 6 | 5 | 4 | 5 | 40 |
| Aikana | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 2 |
| Ainu | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Algic | 0 | 3 | 2 | 2 | 2 | 3 | 1 | 2 | 15 |
| Angan | 3 | 1 | 2 | 3 | 2 | 0 | 2 | 1 | 14 |
| Anson_Bay | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 4 |
| Araucanian | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 0 | 5 |
| Arawakan | 2 | 2 | 2 | 2 | 5 | 2 | 2 | 4 | 21 |
| Arawan | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Athapaskan-Eyak-Tlingit | 0 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 9 |
| Atlantic-Congo | 6 | 6 | 5 | 6 | 6 | 6 | 6 | 6 | 47 |
| Austroasiatic | 4 | 1 | 4 | 3 | 3 | 1 | 2 | 3 | 21 |
| Austronesian | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 6 | 48 |
| Barbacoan | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 5 |
| Basque | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Birri | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Blue_Nile_Mao | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 3 |
| Bogaya | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Boran | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 3 |
| Border | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Bosavi | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 2 | 5 |
| Bulaka_River | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 3 |
| Bunaban | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 3 |
| Burushaski | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 1 | 5 |
| Caddoan | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Cahuapanan | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Cariban | 3 | 3 | 2 | 3 | 0 | 2 | 4 | 1 | 18 |
| Central_Sudanic | 2 | 3 | 3 | 2 | 1 | 2 | 2 | 3 | 18 |
| Chapacuran | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 3 |
| Chibchan | 2 | 2 | 1 | 0 | 0 | 2 | 1 | 1 | 9 |
| Chimakuan | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Chocoan | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Chukotko-Kamchatkan | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 |
| Cochimi-Yuman | 1 | 0 | 1 | 2 | 0 | 1 | 0 | 0 | 5 |
| Comecrudan | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 |
| Daju | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 6 |
| Dibiyaso | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| Dizoid | 0 | 0 | 1 | 1 | 2 | 1 | 0 | 0 | 5 |
| Dogon | 0 | 0 | 2 | 0 | 1 | 0 | 1 | 1 | 5 |
| Doso-Turumsa | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 2 |
| Dravidian | 4 | 3 | 1 | 1 | 1 | 0 | 3 | 1 | 14 |
| East_Bird’s_Head | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 3 |
| East_Strickland | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 3 |
| Eastern_Daly | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 4 |
| Eastern_Trans-Fly | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 3 |
| Eleman | 1 | 1 | 1 | 1 | 0 | 1 | 2 | 2 | 9 |
| Eskimo-Aleut | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 |
| Fasu | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Gaagudju | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Giimbiyu | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Goilalan | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Great_Andamanese | 0 | 0 | 1 | 1 | 0 | 1 | 2 | 1 | 6 |
| Guaicuruan | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Gumuz | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 4 |
| Gunwinyguan | 2 | 1 | 0 | 0 | 1 | 1 | 1 | 0 | 6 |
| Hatam-Mansim | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 2 | 6 |
| Heiban | 0 | 1 | 2 | 2 | 1 | 1 | 2 | 2 | 11 |
| Hibito-Cholon | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Hmong-Mien | 0 | 1 | 1 | 2 | 2 | 1 | 0 | 1 | 8 |
| Huavean | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 3 |
| Huitotoan | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 2 |
| Ijoid | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 3 |
| Inland_Gulf_of_Papua | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 |
| Iroquoian | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 | 4 |
| Itonama | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Iwaidjan_Proper | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 3 |
| Japonic | 0 | 2 | 0 | 1 | 0 | 1 | 2 | 1 | 7 |
| Jivaroan | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 4 |
| Jodi | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Kadugli-Krongo | 2 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 7 |
| Kaki_Ae | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Kakua-Nukak | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Kamsa | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| Kanoe | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Kapauri | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Kartvelian | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 3 |
| Katla-Tima | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | 3 |
| Kawesqar | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Khoe-Kwadi | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 3 |
| Kiowa-Tanoan | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Kiwaian | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 3 |
| Klamath-Modoc | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 3 |
| Kolopom | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Koman | 0 | 2 | 1 | 0 | 0 | 2 | 2 | 1 | 8 |
| Korean | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Kosare | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| Kresh-Aja | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 5 |
| Kujarge | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Kuliak | 0 | 1 | 0 | 1 | 0 | 2 | 0 | 1 | 5 |
| Kunama | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Kuot | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Kusunda | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Kwalean | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| Lakes_Plain | 0 | 1 | 1 | 1 | 1 | 3 | 2 | 1 | 10 |
| Leko | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Lengua-Mascoy | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| Lepki-Murkim | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Limilngan | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 2 |
| Lower_Sepik-Ramu | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 3 |
| Maban | 0 | 2 | 0 | 0 | 0 | 2 | 2 | 0 | 6 |
| Maiduan | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 4 |
| Mande | 2 | 0 | 3 | 2 | 2 | 1 | 1 | 1 | 12 |
| Maningrida | 1 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 5 |
| Manubaran | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Marindic | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 4 |
| Matacoan | 1 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 4 |
| Mayan | 0 | 1 | 1 | 1 | 2 | 2 | 0 | 1 | 8 |
| Mirndi | 0 | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 3 |
| Misumalpan | 0 | 0 | 1 | 3 | 0 | 1 | 0 | 1 | 6 |
| Miwok-Costanoan | 3 | 2 | 1 | 0 | 1 | 0 | 2 | 1 | 10 |
| Mixe-Zoque | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 4 |
| Molala | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 4 |
| Mombum | 0 | 2 | 1 | 0 | 2 | 2 | 0 | 0 | 7 |
| Mongolic | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 3 |
| Morehead-Wasur | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 5 |
| Mpur | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Muniche | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 |
| Muskogean | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 3 |
| Nakh-Daghestanian | 2 | 4 | 0 | 3 | 1 | 3 | 3 | 3 | 19 |
| Nambiquaran | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| Narrow_Talodi | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 4 |
| Natchez | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 0 | 4 |
| Ndu | 0 | 1 | 1 | 0 | 1 | 1 | 0 | 0 | 4 |
| Nilotic | 3 | 1 | 5 | 2 | 3 | 0 | 2 | 3 | 19 |
| Nimboran | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| Nivkh | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| North_Bougainville | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 |
| North_Halmahera | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 4 |
| Northern_Daly | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 4 |
| Nubian | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 3 |
| Nuclear_Torricelli | 2 | 3 | 2 | 2 | 2 | 2 | 3 | 3 | 19 |
| Nuclear_Trans_New_Guinea | 6 | 6 | 6 | 5 | 6 | 5 | 6 | 6 | 46 |
| Nuclear-Macro-Je | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 1 | 5 |
| Nyulnyulan | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 3 |
| Ongota | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| Otomanguean | 4 | 3 | 3 | 5 | 5 | 4 | 1 | 1 | 26 |
| Paez | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| Palaihnihan | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 2 |
| Pama-Nyungan | 2 | 5 | 5 | 4 | 4 | 3 | 4 | 3 | 30 |
| Panoan | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 5 |
| Pauwasi | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| Peba-Yagua | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 3 |
| Puelche | 0 | 0 | 0 | 0 | 1 | 1 | 3 | 0 | 5 |
| Purari | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Quechuan | 2 | 1 | 2 | 0 | 0 | 0 | 2 | 1 | 8 |
| Sahaptian | 1 | 0 | 0 | 1 | 1 | 1 | 1 | 0 | 5 |
| Saharan | 0 | 1 | 1 | 1 | 2 | 1 | 2 | 1 | 9 |
| Salishan | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 0 | 10 |
| Sandawe | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| Sentanic | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| Sepik | 1 | 2 | 2 | 0 | 1 | 1 | 3 | 2 | 12 |
| Sino-Tibetan | 6 | 6 | 4 | 5 | 6 | 3 | 5 | 6 | 41 |
| Siouan | 1 | 1 | 0 | 2 | 1 | 2 | 3 | 0 | 10 |
| Siuslaw | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| Sko | 1 | 1 | 1 | 1 | 0 | 0 | 2 | 0 | 6 |
| Songhay | 1 | 1 | 0 | 0 | 1 | 0 | 2 | 1 | 6 |
| South_Bird’s_Head_Family | 1 | 1 | 1 | 0 | 1 | 1 | 0 | 2 | 7 |
| South_Omotic | 0 | 0 | 0 | 2 | 1 | 1 | 1 | 1 | 6 |
| Southern_Daly | 1 | 1 | 1 | 1 | 0 | 0 | 1 | 1 | 6 |
| Suki-Gogodala | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Surmic | 2 | 1 | 1 | 3 | 2 | 1 | 2 | 2 | 14 |
| Ta-Ne-Omotic | 1 | 1 | 1 | 0 | 1 | 4 | 2 | 1 | 11 |
| Tacanan | 1 | 1 | 1 | 1 | 0 | 1 | 0 | 1 | 6 |
| Tai-Kadai | 3 | 2 | 2 | 4 | 1 | 4 | 1 | 3 | 20 |
| Taiap | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 |
| Teberan | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 3 |
| Ticuna-Yuri | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Timor-Alor-Pantar | 1 | 1 | 1 | 3 | 0 | 2 | 0 | 2 | 10 |
| Timucua | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Tirio | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Totonacan | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 2 | 5 |
| Touo | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 3 |
| Tucanoan | 2 | 3 | 0 | 1 | 2 | 2 | 3 | 1 | 14 |
| Tungusic | 1 | 0 | 3 | 1 | 1 | 1 | 1 | 3 | 11 |
| Tunica | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| Tupian | 2 | 1 | 3 | 0 | 2 | 2 | 2 | 3 | 15 |
| Turama-Kikori | 1 | 0 | 1 | 0 | 1 | 1 | 1 | 1 | 6 |
| Turkic | 3 | 2 | 1 | 1 | 0 | 1 | 2 | 1 | 11 |
| Uralic | 1 | 3 | 1 | 3 | 1 | 2 | 4 | 0 | 15 |
| Uru-Chipaya | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 3 |
| Uto-Aztecan | 2 | 0 | 2 | 1 | 3 | 1 | 2 | 3 | 14 |
| Vilela | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Wagiman | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 2 |
| Wakashan | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| Waorani | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 2 |
| Warao | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| West_Bomberai | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 3 |
| Western_Daly | 0 | 0 | 2 | 1 | 3 | 0 | 0 | 2 | 8 |
| Wintuan | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| Wiru | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| Yamana | 0 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 4 |
| Yana | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 2 |
| Yanomamic | 0 | 0 | 0 | 1 | 1 | 2 | 1 | 1 | 6 |
| Yareban | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
| Yaruro | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Yeli_Dnye | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Yokutsan | 0 | 1 | 1 | 0 | 0 | 1 | 1 | 1 | 5 |
| Yuchi | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Yukaghir | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 2 |
| Yuki-Wappo | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Yurakare | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Zaparoan | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
write.csv(cells, file="Exp1-meanings-by-matching-by-RR.csv", row.names=FALSE, quote=FALSE)
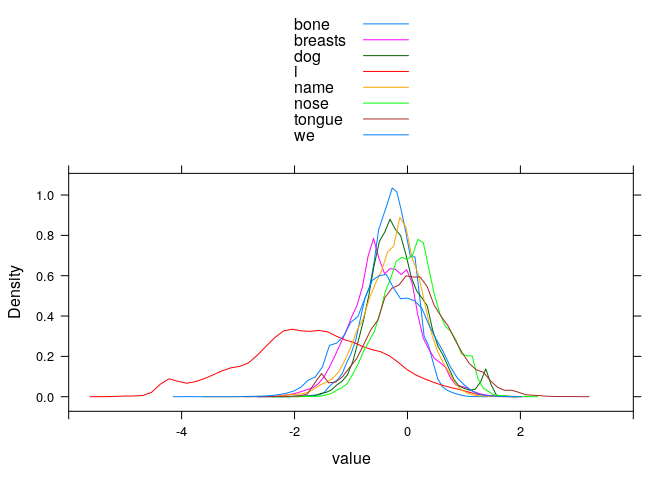
longdata <- reshape2::melt(data, id.vars="Meaning", measure.vars=meaning.columns)
# distribution of RRs per meaning (accurate words only)
#lattice::histogram(~ value | Meaning, type="count", layout=c(4, 2))
lattice::densityplot(~ value, groups=Meaning, subset(longdata, Meaning == variable),
plot.points=FALSE, auto.key=TRUE, xlim=c(-6, 4))
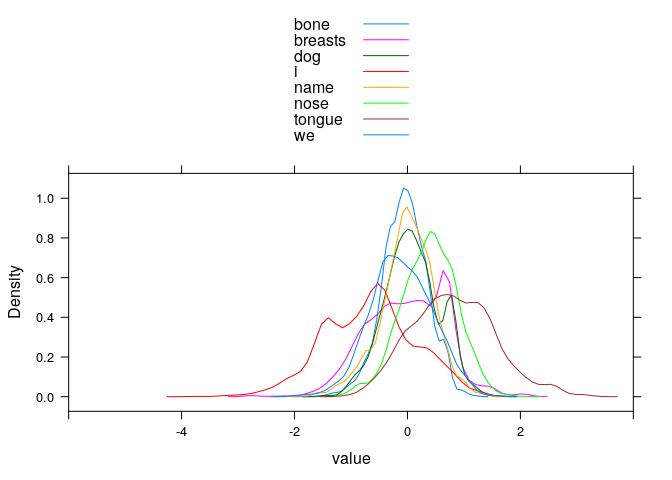
# distribution of RRs per meaning (incorrect words only)
#lattice::histogram(~ value | variable, type="count", layout=c(4, 2))
lattice::densityplot(~ value, groups=variable, subset(longdata, Meaning != variable),
plot.points=FALSE, auto.key=TRUE, xlim=c(-6, 4))